Welcome to the User Manual for Tool 2
This Tool is part of a suite of genome analysis tools that explore bacterial genomes. The suite includes:
| Application | Description |
|---|---|
| The Core | Genome assembly, taxonomic classification, phylogeny, annotation and mass screening. |
| Tool 1 | Mass screening with curated databases. |
| Tool 2 | Mass screening with genes you choose. |
| Tool 3 | Comparison of genomes, and phylogeny. |
| Tool 4 | Primer design: identification of unique stretches of DNA. |
They are built with the pipeline manager Nextflow, and operate within Singularity containers.
The suite was developed as part of a collaborative project between Volac International Ltd. and Cardiff University, partly funded by Innovate UK as part of a knowledge transfer partnership (KTP).
What is Tool 2 for?
Tool 2 screens genome assemblies for specific genes (or any DNA sequence) of your choosing.
What does Tool 2 do?
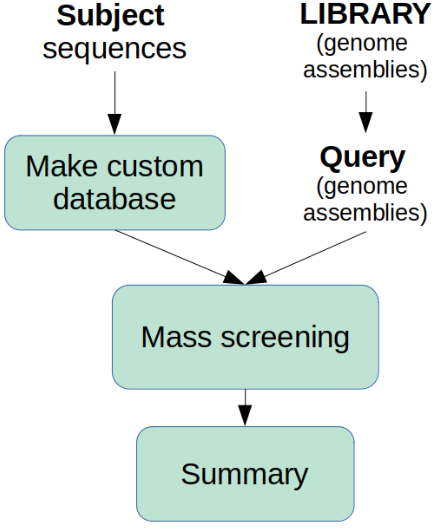
Firstly, Tool 2 takes your custom sequences (subject) and makes a database using BLAST. Secondly, Tool 2 uses this database to screen assembled genomes with the ABRicate application. And thirdly, Tool 2 makes a summary.
Tool 2 screens all genomes in the input_genomes folder. The genome assemblies for all the sequenced strains are saved in the LIBRARY folder.