Using updated software versions
The Tools are functional in their original configuration.
Each Tool is a pipeline that connects many pieces of software. Each piece of software can be exchanged for a different version of the same software. An alternative version could be newer, older, or from a different source.
Each piece of software is housed in a stand-alone unit called a container. Containers are portable and reproducible; they allow the packaging of software and their associated environments (OS, libraries, tools, etc.) within an encapsulated file.
To use a different software version simply specify a different container.
Image Registries
The first thing you need to do is choose the a container.
Biocontainers and StaphB are both a good source of up-to-date and reliable container images for genomic analysis.
Container Images can also be found in:
- Sylabs.io Container Library
- Docker Hub
- Singularity Hub (no longer updated)
- GitHub
For example:
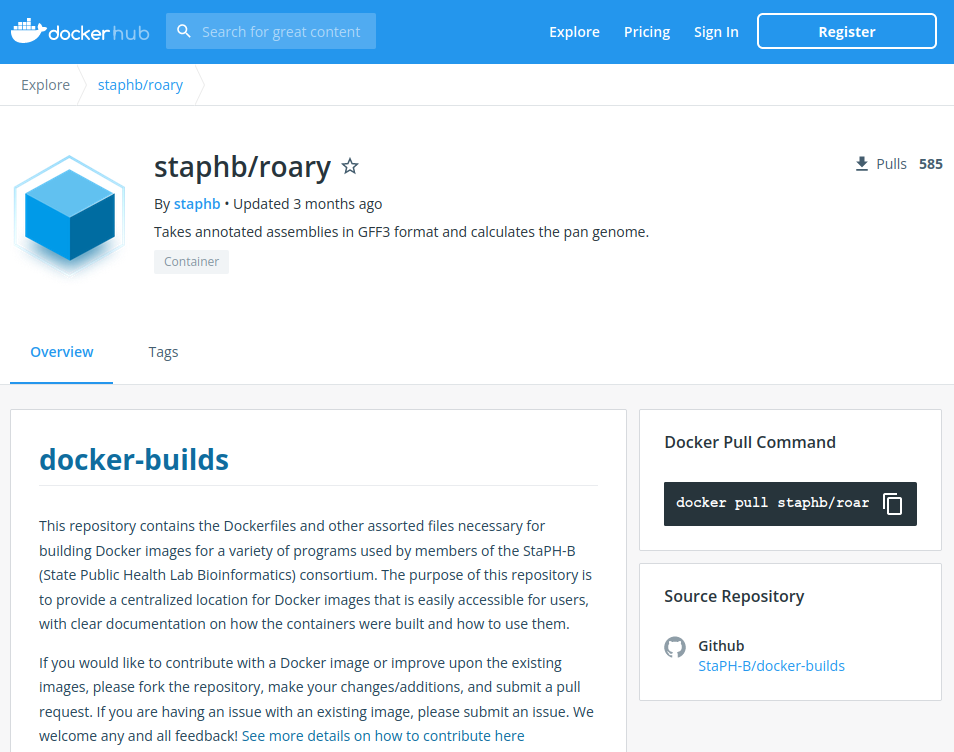
To choose the version of the software tool look in the tab named 'tags'; the different versions are listed.
Specifying alternative software version(s)
The Core, and each Tools all have an individual nextflow.config file. This is where the containers (and consequently, the encapsulated software) is specified.
To edit the nextflow.config file:
- located the relevant
nextflow.configfile - double click to open the file
- update the container address ONLY
- run the pipeline to confirm that the alternative container is functional
For example, in this case, the trimGaloreFastQC process uses the container 'quay.io/biocontainers/trim-galore:0.6.6--0',
process {
withName:trimGaloreFastQC {
container = 'quay.io/biocontainers/trim-galore:0.6.6--0'
}
which can be updated to 'quay.io/biocontainers/trim-galore:0.6.7--hdfd78af_0'.
process {
withName:trimGaloreFastQC {
container = 'quay.io/biocontainers/trim-galore:0.6.7--hdfd78af_0'
}
Important
You must modify all containers that use the same software.
For example, seqtk, seqtkSummary and version_seqtk all use 'quay.io/biocontainers/seqtk:1.3--h5bf99c6_3'. Therefore, change all three.
Original configurations for Core
process {
withName:version_trim_galore {
container = 'quay.io/biocontainers/trim-galore:0.6.7--hdfd78af_0'
}
withName:trimGaloreFastQC {
container = 'quay.io/biocontainers/trim-galore:0.6.7--hdfd78af_0'
}
withName:version_seqtk {
container = 'quay.io/biocontainers/seqtk:1.3--h84994c4_1'
}
withName:seqtk {
container = 'quay.io/biocontainers/seqtk:1.3--h84994c4_1'
}
withName:seqtkSummary {
container = 'quay.io/biocontainers/seqtk:1.3--h84994c4_1'
}
withName:version_shovill {
container = 'quay.io/biocontainers/shovill:1.1.0--hdfd78af_1'
}
withName:shovill {
container = 'quay.io/biocontainers/shovill:1.1.0--hdfd78af_1'
}
withName:version_quast {
container = 'nanozoo/quast:5.0.2--e7f0cfe'
}
withName:quast {
container = 'nanozoo/quast:5.0.2--e7f0cfe'
}
withName:version_prokka {
container = 'quay.io/biocontainers/prokka:1.14.6--pl5321hdfd78af_4'
}
withName:prokka {
container = 'quay.io/biocontainers/prokka:1.14.6--pl5321hdfd78af_4'
}
withName:version_multiQC {
container = 'ewels/multiqc:v1.11'
}
withName:multiQC {
container = 'ewels/multiqc:v1.11'
}
withName:version_abricate {
container = 'nanozoo/abricate:1.0.1--8960147'
}
withName:massScreeningDatabaseDownload {
container = 'nanozoo/abricate:1.0.1--8960147'
}
withName:massScreening {
container = 'nanozoo/abricate:1.0.1--8960147'
}
withName:massScreeningSummary {
container = 'nanozoo/abricate:1.0.1--8960147'
}
withName:version_kraken2 {
container = 'quay.io/biocontainers/kraken2:2.1.2--pl5321h9f5acd7_2'
}
withName:kraken2 {
container = 'quay.io/biocontainers/kraken2:2.1.2--pl5321h9f5acd7_2'
}
withName:kraken2_miniKraken {
container = 'quay.io/biocontainers/kraken2:2.1.2--pl5321h9f5acd7_2'
}
withName:kraken2_QC {
container = 'quay.io/biocontainers/kraken2:2.1.2--pl5321h9f5acd7_2'
}
withName:version_fastANI {
container = 'quay.io/biocontainers/fastani:1.33--h0fdf51a_0'
}
withName:fastANI {
container = 'quay.io/biocontainers/fastani:1.33--h0fdf51a_0'
}
withName:version_fasttree {
container = 'quay.io/biocontainers/fasttree:2.1.10--h779adbc_6'
}
withName:fasttree {
container = 'quay.io/biocontainers/fasttree:2.1.10--h779adbc_6'
}
withName:lsBSR_NEW {
container = 'quay.io/annacprice/extractcoregenome:1.03-r1'
}
}
Original configurations for Tool 1
process {
container = 'nanozoo/abricate:1.0.1--8960147'
}
Original configurations for Tool 2
process {
withName:version {
container = 'nanozoo/abricate:1.0.1--8960147'
}
withName:make_custom_DB {
container = 'ncbi/blast:2.13.0'
}
withName:massScreening {
container = 'nanozoo/abricate:1.0.1--8960147'
}
withName:massScreeningSummary {
container = 'nanozoo/abricate:1.0.1--8960147'
}
}
Original configurations for Tool 3
process {
withName:roary {
container = 'docker://quay.io/biocontainers/roary:3.13.0--pl526h516909a_0'
}
withName:fasttree {
container = 'docker://quay.io/biocontainers/fasttree:2.1.11--h779adbc_0'
}
}
Original configurations for Tool 4
process {
withName:predictORFs {
container = 'quay.io/biocontainers/prodigal:2.6.3--h516909a_2'
}
withName:makeBLASTDatabase {
container = 'ncbi/blast:2.13.0'
}
withName:BLASTGeneSequences {
container = 'ncbi/blast:2.13.0'
}
withName:extractNoHitGenes {
container = 'staphb/samtools:1.15'
}
withName:sortNoHitGenes {
container = 'nanozoo/bbmap:38.86--9ebcbfa'
}
withName:getLongestGenes {
container = 'quay.io/biocontainers/seqtk:1.3--h84994c4_1'
}
}